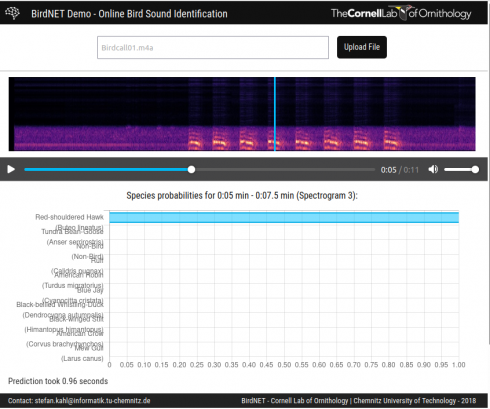
The Cornell Ornithology Lab’s BirdNet lets you upload audio files of bird calls and identifies the birds. I tried it with this file (BirdCall01.m4a) recorded near school, and it identified Red Shouldered Hawks (about 6 seconds in).
Middle and High School … from a Montessori Point of View
The Cornell Ornithology Lab’s BirdNet lets you upload audio files of bird calls and identifies the birds. I tried it with this file (BirdCall01.m4a) recorded near school, and it identified Red Shouldered Hawks (about 6 seconds in).

While teaching chemistry this year, I’ve needed a quick way to write chemical formulas. The fastest thing to do is just write it out flat–sans sub and superscripts–like Na+, or CO2-. But that’s not pretty, and introduces another potential element that could confuse.
I found that you can use Lingojam’s sub and superscript generators to copy and paste the official UTF-8 characters for pretty much all the sub and superscripts I need for chemistry, but that takes a while to do.
So instead, I put together, just in time to write my finals, a little ChemEqn app that uses keyboard shortcuts to quickly substitute in the sub and superscripts as you type.
The method is not perfect, since you still have to decide which comes first when you have both a sub and superscript after an element (O₃²⁻ for example). Another issue is that when I do [Ctrl][Shift][+] and [Ctrl][Shift][-] it zooms into and out of the browser window since we’re using the default zoom shortcuts, but that’s, at least for me, a minor inconvenience.
I do like the app since it still makes for quite readable formulas that can be easily copied and pasted almost anywhere without messing up since it only uses UTF-8 characters that are pretty standard across the web (and most computer programs).
Windyty shows beautiful animations of the current wind patterns around the world.
Calculate the molar mass of a molecule:
The notation for the chemical formula is a little funky: you put the element symbol and then the number of atoms separated by a colon; each element/number of atoms pair are separated by commas, so sodium chloride (NaCl) would be “Na:1,Cl:1“.
This will have to do until I can write something to parse the regular chemical formula notation.
On the plus side, you can link to a specific molecular mass calculation by adding the formula to the url. So magnesium chloride (MgCl2) can be found with this url:
https://soriki.com/js/chem/chem_db/molecular_mass.html?formula=Mg:1,Cl:2
A couple of my students asked for worksheets to practice drawing atoms and electron shells. I updated the Atom Builder app to make sure it works and to make the app embedable.
So now I can ask a student to draw 23Na+ then show the what they should get:
Draw diagrams of the following atoms, showing the number of neutrons, protons, and electrons in shells. See the example above.
I guess the next step is to adapt the app so you can hide the element symbol so student have to figure what element based on the diagram.
My students (almost) created an unmanned robotic drone for delivering messages to the main office.
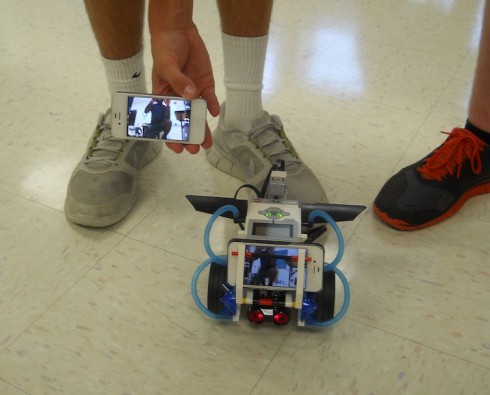
The robotics students have put together their first robots and are trying out the software for making them move. Lego has a new app that allows you to use your iPhone or iPad as a remote control, and my kids love it. They’ve got a robot fighting competition going on using them as remotes. I, however, wanting to push them into actually programming the robots for autonomous motion, told them I wanted them to get the robot to deliver a message to the front office without them following it through the building.
Well, this was their solution:
They attached an iPhone to one of their robot with the camera facing out. They then connected made a video call to that phone using another phone and the FaceTime app. Since the two phones were busy communicating with each other, the robot was operated using the wireless, Bluetooth connection via someone else’s iPad.
It worked. More or less. The robot lost the Bluetooth connection at the end of the hall, so the robot could not make it all the way to the office.
So now they’re actually trying to write a program. Although I finally got them to do what I wanted, I have to say I was impressed with their innovative use of technology.
Try it. You can change the order and coefficients of the polynomial. The default is the second order polynomial: y = x2.
I originally started putting together this interactive polynomial app to use in demonstrating numerical integration, however it’s a quite useful thing on its own. In fact, I’ve finally figured out how to do iframes, which means that the app is embeddable, so you can use it directly off the Muddle (if you want to put it on your own website you can get the embed code).
This app is a rewritten version of the parabola code, but it uses kineticjs instead of just HTML5 canvases. As a result, it should be much easier to adapt to make it touch/mouse interactive.
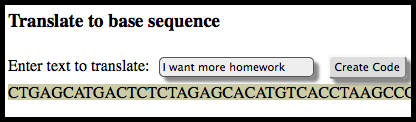
I created this little DNA Writer webpage after seeing the article on scientists recording one of Shakespeare’s sonnets on DNA, I was inspired to put together something similar as an assignment for my middle-school science class to demonstrate how DNA records information. With the website to do quick translations for me, I’ll give each student the translation table and a simple message in DNA code and have them figure out the message.
Update: I’ve adapted the code to add a two to five letter sequence of non-coding DNA to the beginning and end of the message code. There’s also start and stop code as well.
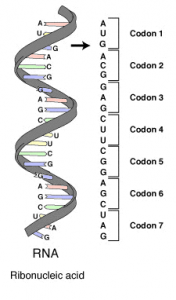
The DNA Writer code uses a simple look-up table where each letter in the English alphabet is assigned a unique three letter nucleotide code. The three letters are chosen from the letters of the DNA bases – AGCT – similar to the way codons are organized in mRNA. Any unknown characters or punctuation are ignored.
Also, with a little tweaking, I think I can adapt this assignment to show how random mutation can be introduced into DNA sequences during transcription. Maybe break the class into groups of 4, give the first student a message as a nucleotide sequence have them copy and pass it on to the next student and so on. If I structure this as a race between the groups, then someone’s bound to introduce some errors, so when they translate the final code back into English they should see how the random mutation affected their code.
UPDATE: Non-Coding (junk) DNA: I’ve updated the code so that you have the option of adding a short (2-5 character) string of non-coding DNA to the beginning and end of each sequence.
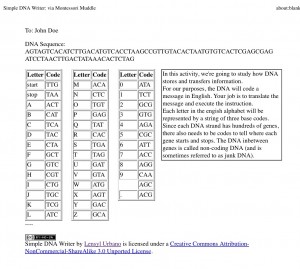
UPDATE 2: Personalized and Printable output: Since I’m using the DNA writer to give each student a personalized message, I’ve created a button that gives “Printer Friendly Output” which will produce an individualized page with the code, the translation table, and some information on how it works, so I can print off individualized assignments more easily.
UPDATE 3: You can now get a color coded version of the sequence.
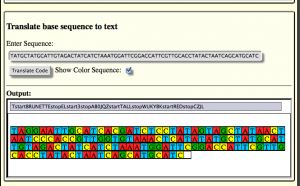
Update 4: Now you can embed the nucleobase color patterns into other websites. Like so:
DNA Writer A: https://earthsciweb.org/js/bio/dna-writerA/
In constructing the codon-to-english conversion table I had to decide if I wanted to go with the standard coding (e.g. letting GTC which codes for alanine represent A) or make up a random encoding.
I opted for the random approach for a number of reasons, but the primary one was that multiple codons can code for the same amino acid. GCT, GCC, GCA, and GCG all code for alanine. This would not necessarily be a problem, except that if we respect all of the multiple encodings, we run out of codons to represent things like numbers and punctuation. A secondary reason is that U is used to represent the 21st amino acid, selenocysteine, but its codon is the same as the stop codon (Croat, 2012) and its addition to the protein chain depends on not just a single codon in the sequence.
I’ve created a hybrid option: dnaWriterA which respects the standard lettering as much as possible (based off of the inverse DNA codon table on Wikipedia). In the table below, the bolded sequences are the ones that have been reassigned.
| Letter/code | Amino acid | Codon | |||||
| start | ATG | ||||||
| stop | TAA | ||||||
| space (” “) | GCA | ||||||
| . | GGA | ||||||
| A | Ala | GCT | GCC | GCA | GCG | ||
| B | Asn or Asp | AAC | |||||
| C | Cys | TGT | TGC | ||||
| D | Asp | GAT | GAC | ||||
| E | Glu | GAA | GAG | ||||
| F | Phe | TTT | TTC | ||||
| G | Gly | GGT | GGC | GGA | GGG | ||
| H | His | CAT | CAC | ||||
| I | Ile | ATT | ATC | ATA | |||
| J | TTG | ||||||
| K | Lys | AAA | |||||
| L | Leu | CTT | CTC | CTA | CTG | TTA | TTG |
| M | Met | ATG | |||||
| N | Asn | AAT | AAC | ||||
| O | AGG | ||||||
| P | Pro | CCT | CCC | CCA | CCG | ||
| Q | Gln | CAA | CAG | ||||
| R | Arg | CGT | CGC | CGA | CGG | AGA | AGG |
| S | Ser | TCT | TCC | TCA | TCG | AGT | AGC |
| T | Thr | ACT | ACC | ACA | ACG | ||
| U | AGA | ||||||
| V | Val | GTT | GTC | GTA | GTG | ||
| W | Trp | TGG | |||||
| X | AGC | ||||||
| Y | Tyr | TAT | TAC | ||||
| Z | Gln or Glu | CAA | CAG | GAA | GAG | ||
| 0 | AGT | ||||||
| 1 | GCG | ||||||
| 2 | GGG | ||||||
| 3 | CTG | ||||||
| 4 | CCG | ||||||
| 5 | CGG | ||||||
| 6 | TCG | ||||||
| 7 | ACG | ||||||
| 8 | GTG | ||||||
| 9 | GAG |
I’ve also posted the code to GitHub: https://github.com/lurbano/dnaWriterA with instructions on how to adapt the sequence.