I need some students to try this at school. Muscle fibers that contract on heating sounds like a great way to open and close vents for air circulation (in the chicken coop to start with).
Author: Lensyl Urbano
Teaching Creativity
Tina Seelig teaches creativity and innovation at Stanford. She has a new book out on the topic called Insight Out. She answered questions about the book recently and gave this answer (I’ve reformatted it into bullet points for clarity):
The model I present in Insight Out describes a series of steps from ideas to actions. Each step requires more effort than the one before.
- Imagination requires engaging and envisioning what might be different.
- Creativity is applying your imagination to solve a problem. This requires motivation and experimentation.
- Innovation is applying creativity to come up with unique solutions. This requires focus and reframing. And,
- entrepreneurship is applying innovations to bring the to the world. This requires persistence and inspiring others.
–Tina Seelig on Reddit.com.
In response to a question about testing students:
- I don’t test my students. In fact, I tell them to never ask about their course grade. 🙂
I want the students to be internally motivated. I tell them that I expect them to put as much work into the course as I do, and that they should “never miss an opportunity to be fabulous.” Guess what? It works! They are waiting for someone to give them this freedom to tap into their own motivation, not respond to an external motivation.
Our Microbial Symbionts
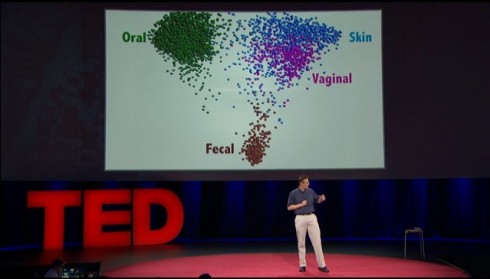
Sometimes I ask my students if we’re not just giant mechs for our microbial symbionts. After all, they outnumber us by about 10 to 1–in our own bodies. Rob Knight’s TED talk stokes my curiosity.
Planting Probabilities
The Gardening Department of our Student-Run-Business sowed seeds in little coconut husk pellets. The question was: how many seeds should we plant per pellet.

Since we’ll only let one seedling grow per pellet, and cull the rest, the more seeds we plant per pellet, the fewer plants we’ll end up with. On the other hand, the fewer seeds we plant (per pellet) the greater the chance that nothing will grow in a particular pellet, and we’ll be down a few plants as well. So we need to think about the probabilities.
Fortunately, I’d planted a some tomato seeds a couple weeks ago that we could use for a test case. Of the 30 seeds I planted, only 20 sprouted, giving a 2/3 probability that any given seed would grow:
![]()
So if we plant one seed per pellet in 10 pellets then in all probability, only two thirds will grow (that’s about 7 out of 10).
What if instead, we planted two seeds per pellet. What’s the probability that at least one will grow. This turns out to be a somewhat tricky problem–as we will see–so let’s set up a table of all the possible outcomes:
| Seed 1 | Seed 2 |
|---|---|
| grow | grow |
| grow | not grow |
| not grow | grow |
| not grow | not grow |
Now, if the probability of a seed growing is 2/3 then the probability of one not growing is 1/3:
![]()
So let’s add this to the table:
| Seed 1 | Seed 2 |
|---|---|
| grow (2/3) | grow (2/3) |
| grow (2/3) | not grow (1/3) |
| not grow (1/3) | grow (2/3) |
| not grow (1/3) | not grow (1/3) |
Now let’s combine the probabilities. Consider the probability of both seeds growing, as in the first row in the table. To calculate the chances of that happening we multiply the probabilities:
![]()
Indeed, we use the ∩ symbol to indicate “and”, so we can rewrite the previous statement as:
![]()
And we can add a new column to the table giving the probability that each row will occur by multiplying the individual probabilities:
| Seed 1 | Seed 2 | And (∩) |
|---|---|---|
| grow (2/3) | grow (2/3) | 4/9 |
| grow (2/3) | not grow (1/3) | 2/9 |
| not grow (1/3) | grow (2/3) | 2/9 |
| not grow (1/3) | not grow (1/3) | 1/9 |
Notice, however, that the two middle outcomes (that one seed grows and the other fails) are identical, so we can say that the probability that only one seed grows will be the probability that the second row happens or that the third row happens:
![]()
When we “or” probabilities we add them together (and we use the symbol ∪) so:
![]()
You’ll also note that the probability that neither seed grows is equal to the probability that seed one does not grow and seed 2 does not grow:
![]()
So we can summarize our possible outcomes a bit by saying:
| Outcome | Probability |
|---|---|
| both seeds grow | 4/9 |
| only one seed grows | 4/9 |
| neither seed grows | 1/9 |
What you can see here, is that the probability that at least one seed grows is the probability that both seeds grow plus the probability that only one seed grows, which is 8/9 (we’re using the “or” operation here again).
In fact, you can calculate this probability by simply taking the opposite probability that neither seeds grow:
![]()
Generalizing a bit, we see that for any number of seeds, the probability that none will grow is the multiplication of individual probability that one seed will not grow:
Probability that no seeds will grow
| Number of seeds | Probability they wont grow | |
|---|---|---|
| 1 | 1/3 | (1/3)1 |
| 2 | (1/3)×(1/3) = 1/9 | (1/3)2 |
| 3 | (1/3)×(1/3)×(1/3) = 1/27 | (1/3)3 |
| n | (1/3)×(1/3)×(1/3)×… | (1/3)n |
So to summarize, the probability that at least one plant will grow, if we plant n seeds is:
![]()
which is:
![]()
Which is something we may have seen before: What are the odds?
Finally to answer our question: how many seeds we should plant, we need to decide how high a probability we need of success:
Probability that at least one seed will grow
| Number of seeds | Probability that at least one seed will grow | % |
|---|---|---|
| 1 | 2/3 | 67% |
| 2 | 8/9 | 89% |
| 3 | 26/27 | 96% |
| 4 | 80/81 | 99% |
| n | 1-(1/3)n |

How to Write a Web Page from Scratch
<html> <head> </head> <body> </body> </html>
I gave a quick introduction to writing HTML web pages over the last interim. The basic outline of a web page can be seen above (but there’s no content in there, so if you tried this file in a web browser you’d get a blank page). Everything is set between tags. The open html tag (<html>) starts the document, and the close html tag (</html>) ends it.
The head section (<head>) holds the information needed to set up the page, while the body section (<body>) holds the stuff that’s actually on the page.
For example, if I wanted to create a heading and a paragraph of text, I would just put it in the body, like so:
<html> <head> </head> <body> <h1>My Heading Here</h1> This is my text. </body> </html>
Note that the heading is placed between h1 tags. And the result should look like this:

Now save this file as an html file, which means save it as a plain text file using a .html extension (e.g. test.html) and open the file in a browser.
Note on Editors: I’ve found that the best way to do this is by using a simple coding editor. I recommend my students use GEdit on Linux, Smultron (free version here) or GEdit on Mac, and Notepad++ on Windows. You don’t want to use something like Microsoft Word, because complex word processing software like it, LiberOffice and Pages need to save a lot of other information about the files as well (like the font style, who created the file etc.).
CSS Styling
But we typically don’t want something so plain, so we’ll style our heading using CSS. Usually, we’ll want all of our headings to be the same so we set up a style to make all of our headings blue. So we’ll add a styling section to our header like so:
<html>
<head>
<style type="text/css">
h1 { color: blue; }
</style>
</head>
<body>
<h1>My Heading Here</h1>
This is my text.
</body>
</html>
Which gives:

There are a few named colors like blue, but if you want more freedom, you can use the hexadecimal color codes like “#A3319F” to give a kind of purple color (hexadecimals are numbers in base 16).
DIV’s to Divide
Finally, we’ll often want pages with separate blocks of information, so let’s make a little column for our heading and paragraph. We’ll make the background yellowish, put a border around it, and give it a set width.
To do this we’ll place our heading and paragraph into a DIV tag, which does not do anything but encapsulate the part of the web page we want into sections.
<html>
<head>
<style type="text/css">
h1 { color: blue; }
</style>
</head>
<body>
<div id="section1">
<h1>My Heading Here</h1>
This is my text.
</div>
</body>
</html>
Note that I’ve given the div tag an id (“section1), this is so that I can refer to that section only in the styling section using “#section”:
<html>
<head>
<style type="text/css">
h1 { color: blue; }
#section1 {
background-color: #A2A82D;
border: solid black 2px;
width: 400px;
}
</style>
</head>
<body>
<div id="section1">
<h1>My Heading Here</h1>
This is my text.
</div>
</body>
</html>
Unleashed
With that introduction, I set students loose to make their own web pages. They had to make two page, both linking to the other, and one of them being an “About Me” page.
There are lots of places on line to find out what HTML tags and CSS styles are available and how to use them, so my goal was to introduce students to the language they needed to know.
One issue that came up was the use of copyrighted images. Current adolescents see everything online as part of a sharing culture–most of my students for this lesson had Pintrest accounts–and it took some explanation for them to understand why they should not use that cute gif of a bunny eating a slice of pizza without getting permission from the author (or at least finding out if the artwork was free to use).
Finally, I did do a quick intro on how to using JavaScript (with Jquery) to make their pages more interactive, but given that we only had two days for this project, that was pushing things a little too far.
Paper Butterflies
Dance Lesson

This interim focuses on our global community. As a result, one of our Chinese students gave a lesson on a traditional dance. I think I made a good start at learning the twirls, but we did not have time to get to the jumps.

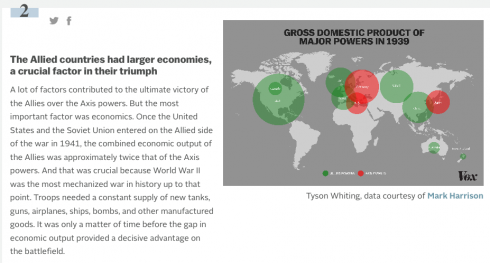
Maps that Explain …
Vox does some great articles using maps to explain things like World War II (which is always a popular middle/high school topic for history papers). What the site does best, is that it accompanies each map with an explanation of what was going on. This results in a set of really interesting vignettes that stoke the curiosity bunkers.